Survival Time Prediction of Metastatic Melanoma Patients by Computed Tomography using Convolutional Neural Networks
Master Thesis (At EnCoV-TGI, Institut Pascal, Universite Clermont Auvergne, France, August 2020)
Metastatic melanoma is a fatal disease with a poor prognosis and rapid systemic spread. Follow-up and study are imperative, especially within the early periods after diagnosis as the expected cure is seldom obtained after surgical excision and with adjuvant therapy. Typically, computed tomography (CT) scan contains a huge sum of data that ought to be completely analyzed and assessed by the radiologist or other healthcare proficient in a brief time. In this case, the CAD framework can be of extraordinary offer assistance as a moment supposition for experts. In spite of the fact that the CAD framework may play an imperative part in the analysis, small research has been published on the survival time prediction of metastatic melanoma based on the CAD system.
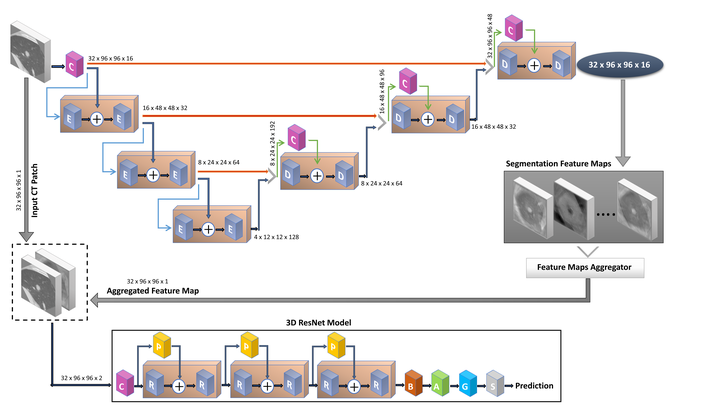
Our objective is to study the prediction of the survival time of patients with metastatic melanoma in terms of 1-year survival as a binary classification. Dataset used in this study contains CTs of 71 patients with metastatic melanoma who are studied at Universite Clermont Auvergne Hospital. The number of lesions per patient varies from 1 to 11. To reach the objective, the survival time is anticipated using the accessible CT data as input of a 3D CNN. In this category, survival time is anticipated using full CT volumes and also from extracted 3D CT patches containing lesion regions. Here, the patches are extracted given the ground truth masks of the lesions. Moreover, segmentation of lesions coming from different organs is performed using two different 3D CNNs to examine the prediction of survival time based on newly extracted 3D CT patches. These new patches are extracted using our anticipated segmentation predicted masks of lesions. As the final test, it is also inspected whether aggregated deep segmentation feature map can help to predict survival time being an extra input channel to the CT data for 3D CNN or not.
Our study shows that using aggregated deep segmentation feature map as an extra input channel to CT data comes about in superior performance in survival time prediction compared to using as it were only 3D CT patches as input. In expansion, the prediction of survival time based on newly extracted 3D CT patches coming from our segmentation predicted masks is similar to the survival prediction using the 3D CT patches coming from ground truth masks.
Further investigation of this study can be the addition of radiomic features with aggregated deep segmentation feature map as additional input to CT data besides experimenting on bigger dataset. Including clinical data such as age, sex, etc. can as well play a vital role.